The marker can be labeled radioactively with T4 Polynucleotide Kinase. The restriction enzymes which we used in this laboratory are EcoRI HindIII and BamHI and their sequences are as follows with the cut site indicated by the arrow.

DNA fragments contain various sticky ends depending on the restriction enzyme used for preparation of the marker.

Hind 3 restriction enzyme. Perform restriction enzyme digestion with reliable restriction endonucleases. In this case the species Haemophilus influenza strain D third isolated enzyme from that strain. HindIII has a High Fidelity version HindIII-HF NEB R3104.
High-Fidelity HF restriction enzymes have the same specificity as native enzymes but have been engineered for significantly reduced star activity and performance in a single buffer CutSmart Buffer. So the correct option is Option B. High Fidelity HF Restriction Enzymes have 100 activity in CutSmart Buffer.
In 1970 Hamilton O. The resulting 7 fragments range from 564 base pairs to 2313 kilobases in size. Welcome to RestrictionMapper - on line restriction mapping the easy way.
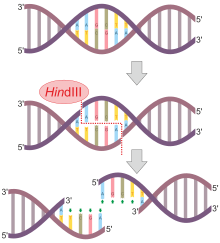
HindIII pronounced Hin D Three is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg 2 via hydrolysis. Coli strain R first. Restriction endonucleases in DNA sequences.
All HF-restriction enzymes come with Gel Loading Dye Purple 6X. Restriction enzyme names tell you exactly were they are from. L cut with Eco RI l cut with Hind III.
65 C for 15 minutes Application HindIII a restriction endonuclease is used in molecular biology applications to cleave DNA at the recognition site 5-AAGCTT-3 to generate DNA fragments with cohesive 5-ends. Restriction fragment length polymorphism analysis also was employed by Waters and Schaal 1991 who could not detect any within- or between-population variation in more than 150 restriction enzyme sites in the chloroplast genome of two populations of Torrey pine separated by 280 km for at least 8000 years. Type-2 restriction enzyme HindIII EC.
The DNA fragments of CFP and YFP are amplified from the commercial vectors pECFP-C1 and pEYFP-C1. Recognition site are GA for EcoRI and AA for HINDIII in 5 3 direction in the sequence to produce sticky ends. HF enzymes also exhibit dramatically reduced star activity.
Single-buffer simplicity means more straightforward and streamlined sample processing. Smith Thomas Kelly and Kent Wilcox isolated and characterized the first type II restriction enzyme Hind II from the bacterium Haemophilus influenzae. 1060AH for complete product documentation and resources.
Hind III High Concentration 50000 Units. DNA Restriction Enzymes from Takara such as HindIII are high-quality. The restriction enzyme Hindlll is a type II site specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae Rd that cleaves the DNA palindromic sequence AAGCTT in the presence of cofactor.
2-10-fold Hind III overdigestion of 1 mg l DNA substrate results in 100 cutting Heat inactivation. 564 2027 2322 4361 6557 9416 23130 bp. Alternatively and it can be labeled radioactively or non-radioactively with Klenow Fragment or Klenow Fragment exo- with the fill-in reaction see Resources for Protocols.
Full-length LC3B or GATE-16 is first cloned into pET-28a between the BamHI and HindIII restriction sites. Hin dIII restrictions process results in formation of overhanging palindromic sticky ends. Thermo Scientific HindIII restriction enzyme recognizes AAGCTT sites and cuts best at 37C in R buffer.
Type II restriction enzyme HindIII Gene names i. Maps sites for restriction enzymes aka. The restriction enzymes studied by Arber and Meselson were type I restriction enzymes which cleave DNA randomly away from the recognition site.
High-Fidelity HF restriction enzymes have the same specificity as native enzymes but have been engineered for significantly reduced star activity and performance in a single buffer CutSmart BufferAll HF-restriction enzymes come with Gel Loading Dye Purple 6XEnjoy the enhanced performance and added value of our engineered enzymes at the same price as the native enzyme. See Reaction Conditions for Restriction Enzymes for a table of enzyme activity conditions for double digestion and heat inactivation for this and other restriction enzymes. Also available as a FastDigest enzyme for rapid DNA digestion.
Lambda HindIII Ladder The l HindIII DNA Ladder is prepared by restriction digestion of phage Lambda DNA to completion with HindIII followed by heat inactivation of the enzyme. Please refer to Cat. Also does virtual digestion.
Http Himedialabs Com Td Htbm015 Pdf
 Activity 3 Restriction Enzyme Analysis
Activity 3 Restriction Enzyme Analysis
 Activity 3 Restriction Enzyme Analysis
Activity 3 Restriction Enzyme Analysis
 Restriction Site Positions And Functions Sigma Aldrich
Restriction Site Positions And Functions Sigma Aldrich
 Problem Solving Test Restriction Endonuclease Mapping Szeberenyi 2011 Biochemistry And Molecular Biology Education Wiley Online Library
Problem Solving Test Restriction Endonuclease Mapping Szeberenyi 2011 Biochemistry And Molecular Biology Education Wiley Online Library
 Solved A Circular 21 Kb Piece Of Dna Is Digested With The Chegg Com
Solved A Circular 21 Kb Piece Of Dna Is Digested With The Chegg Com
 1 12 Restriction Digest With Gel Electrophorisis Biology Libretexts
1 12 Restriction Digest With Gel Electrophorisis Biology Libretexts